A gentle introduction to text mining using R
Preamble
This article is based on my exploration of the basic text mining capabilities of R, the open source statistical software. It is intended primarily as a tutorial for novices in text mining as well as R. However, unlike conventional tutorials, I spend a good bit of time setting the context by describing the problem that led me to text mining and thence to R. I also talk about the limitations of the techniques I describe, and point out directions for further exploration for those who are interested. Indeed, I’ll likely explore some of these myself in future articles.
If you have no time and /or wish to cut to the chase, please go straight to the section entitled, Preliminaries – installing R and RStudio. If you have already installed R and have worked with it, you may want to stop reading as I doubt there’s anything I can tell you that you don’t already know 🙂
A couple of warnings are in order before we proceed. R and the text mining options we explore below are open source software. Version differences in open source can be significant and are not always documented in a way that corporate IT types are used to. Indeed, I was tripped up by such differences in an earlier version of this article (now revised). So, just for the record, the examples below were run on version 3.2.0 of R and version 0.6-1 of the tm (text mining) package for R. A second point follows from this: as is evident from its version number, the tm package is still in the early stages of its evolution. As a result – and we will see this below – things do not always work as advertised. So assume nothing, and inspect the results in detail at every step. Be warned that I do not always do this below, as my aim is introduction rather than accuracy.
Background and motivation
Traditional data analysis is based on the relational model in which data is stored in tables. Within tables, data is stored in rows – each row representing a single record of an entity of interest (such as a customer or an account). The columns represent attributes of the entity. For example, the customer table might consist of columns such as name, street address, city, postcode, telephone number . Typically these are defined upfront, when the data model is created. It is possible to add columns after the fact, but this tends to be messy because one also has to update existing rows with information pertaining to the added attribute.
As long as one asks for information that is based only on existing attributes – an example being, “give me a list of customers based in Sydney” – a database analyst can use Structured Query Language (the defacto language of relational databases ) to get an answer. A problem arises, however, if one asks for information that is based on attributes that are not included in the database. An example in the above case would be: “give me a list of customers who have made a complaint in the last twelve months.”
As a result of the above, many data modelers will include a “catch-all” free text column that can be used to capture additional information in an ad-hoc way. As one might imagine, this column will often end up containing several lines, or even paragraphs of text that are near impossible to analyse with the tools available in relational databases.
(Note: for completeness I should add that most database vendors have incorporated text mining capabilities into their products. Indeed, many of them now include R…which is another good reason to learn it.)
My story
Over the last few months, when time permits, I’ve been doing an in-depth exploration of the data captured by my organisation’s IT service management tool. Such tools capture all support tickets that are logged, and track their progress until they are closed. As it turns out, there are a number of cases where calls are logged against categories that are too broad to be useful – the infamous catch-all category called “Unknown.” In such cases, much of the important information is captured in a free text column, which is difficult to analyse unless one knows what one is looking for. The problem I was grappling with was to identify patterns and hence define sub-categories that would enable support staff to categorise these calls meaningfully.
One way to do this is to guess what the sub-categories might be…and one can sometimes make pretty good guesses if one knows the data well enough. In general, however, guessing is a terrible strategy because one does not know what one does not know. The only sensible way to extract subcategories is to analyse the content of the free text column systematically. This is a classic text mining problem.
Now, I knew a bit about the theory of text mining, but had little practical experience with it. So the logical place for me to start was to look for a suitable text mining tool. Our vendor (who shall remain unnamed) has a “Rolls-Royce” statistical tool that has a good text mining add-on. We don’t have licenses for the tool, but the vendor was willing to give us a trial license for a few months…with the understanding that this was on an intent-to-purchase basis.
I therefore started looking at open source options. While doing so, I stumbled on an interesting paper by Ingo Feinerer that describes a text mining framework for the R environment. Now, I knew about R, and was vaguely aware that it offered text mining capabilities, but I’d not looked into the details. Anyway, I started reading the paper…and kept going until I finished.
As I read, I realised that this could be the answer to my problems. Even better, it would not require me trade in assorted limbs for a license.
I decided to give it a go.
Preliminaries – installing R and RStudio
R can be downloaded from the R Project website. There is a Windows version available, which installed painlessly on my laptop. Commonly encountered installation issues are answered in the (very helpful) R for Windows FAQ.
RStudio is an integrated development environment (IDE) for R. There is a commercial version of the product, but there is also a free open source version. In what follows, I’ve used the free version. Like R, RStudio installs painlessly and also detects your R installation.
RStudio has the following panels:
- A script editor in which you can create R scripts (top left). You can also open a new script editor window by going to File > New File > RScript.
- The console where you can execute R commands/scripts (bottom left)
- Environment and history (top right)
- Files in the current working directory, installed R packages, plots and a help display screen (bottom right).
Check out this short video for a quick introduction to RStudio.
You can access help anytime (within both R and RStudio) by typing a question mark before a command. Exercise: try this by typing ?getwd() and ?setwd() in the console.
I should reiterate that the installation process for both products was seriously simple…and seriously impressive. “Rolls-Royce” business intelligence vendors could take a lesson from that…in addition to taking a long hard look at the ridiculous prices they charge.
There is another small step before we move on to the fun stuff. Text mining and certain plotting packages are not installed by default so one has to install them manually The relevant packages are:
- tm – the text mining package (see documentation). Also check out this excellent introductory article on tm.
- SnowballC – required for stemming (explained below).
- ggplot2 – plotting capabilities (see documentation)
- wordcloud – which is self-explanatory (see documentation) .
(Warning for Windows users: R is case-sensitive so Wordcloud != wordcloud)
The simplest way to install packages is to use RStudio’s built in capabilities (go to Tools > Install Packages in the menu). If you’re working on Windows 7 or 8, you might run into a permissions issue when installing packages. If you do, you might find this advice from the R for Windows FAQ helpful.
Preliminaries – The example dataset
The data I had from our service management tool isn’t the best dataset to learn with as it is quite messy. But then, I have a reasonable data source in my virtual backyard: this blog. To this end, I converted all posts I’ve written since Dec 2013 into plain text form (30 posts in all). You can download the zip file of these here .
I suggest you create a new folder called – called, say, TextMining – and unzip the files in that folder.
That done, we’re good to start…
Preliminaries – Basic Navigation
A few things to note before we proceed:
- In what follows, I enter the commands directly in the console. However, here’s a little RStudio tip that you may want to consider: you can enter an R command or code fragment in the script editor and then hit Ctrl-Enter (i.e. hit the Enter key while holding down the Control key) to copy the line to the console. This will enable you to save the script as you go along.
- In the code snippets below, the functions / commands to be typed in the R console are in blue font. The output is in black. I will also denote references to functions / commands in the body of the article by italicising them as in “setwd()”. Be aware that I’ve omitted the command prompt “>” in the code snippets below!
- It is best not to cut-n-paste commands directly from the article as quotes are sometimes not rendered correctly. A text file of all the code in this article is available here.
The > prompt in the RStudio console indicates that R is ready to process commands.
To see the current working directory type in getwd() and hit return. You’ll see something like:
[1] “C:/Users/Documents”
The exact output will of course depend on your working directory. Note the forward slashes in the path. This is because of R’s Unix heritage (backslash is an escape character in R.). So, here’s how would change the working directory to C:\Users:
You can now use getwd()to check that setwd() has done what it should.
[1]”C:/Users”
I won’t say much more here about R as I want to get on with the main business of the article. Check out this very short introduction to R for a quick crash course.
Loading data into R
Start RStudio and open the TextMining project you created earlier.
The next step is to load the tm package as this is not loaded by default. This is done using the library() function like so:
Dependent packages are loaded automatically – in this case the dependency is on the NLP (natural language processing) package.
Next, we need to create a collection of documents (technically referred to as a Corpus) in the R environment. This basically involves loading the files created in the TextMining folder into a Corpus object. The tm package provides the Corpus() function to do this. There are several ways to create a Corpus (check out the online help using ? as explained earlier). In a nutshell, the Corpus() function can read from various sources including a directory. That’s the option we’ll use:
docs <- Corpus(DirSource(“C:/Users/Kailash/Documents/TextMining”))
At the risk of stating the obvious, you will need to tailor this path as appropriate.
A couple of things to note in the above. Any line that starts with a # is a comment, and the “<-“ tells R to assign the result of the command on the right hand side to the variable on the left hand side. In this case the Corpus object created is stored in a variable called docs. One can also use the equals sign (=) for assignment if one wants to.
Type in docs to see some information about the newly created corpus:
Metadata: corpus specific: 0, document level (indexed): 0
Content: documents: 30
The summary() function gives more details, including a complete listing of files…but it isn’t particularly enlightening. Instead, we’ll examine a particular document in the corpus.
writeLines(as.character(docs[[30]]))
Which prints the entire content of 30th document in the corpus to the console.
Pre-processing
Data cleansing, though tedious, is perhaps the most important step in text analysis. As we will see, dirty data can play havoc with the results. Furthermore, as we will also see, data cleaning is invariably an iterative process as there are always problems that are overlooked the first time around.
The tm package offers a number of transformations that ease the tedium of cleaning data. To see the available transformations type getTransformations() at the R prompt:
> getTransformations()
[1] “removeNumbers” “removePunctuation” “removeWords” “stemDocument” “stripWhitespace”
Most of these are self-explanatory. I’ll explain those that aren’t as we go along.
There are a few preliminary clean-up steps we need to do before we use these powerful transformations. If you inspect some documents in the corpus (and you know how to do that now), you will notice that I have some quirks in my writing. For example, I often use colons and hyphens without spaces between the words separated by them. Using the removePunctuation transform without fixing this will cause the two words on either side of the symbols to be combined. Clearly, we need to fix this prior to using the transformations.
To fix the above, one has to create a custom transformation. The tm package provides the ability to do this via the content_transformer function. This function takes a function as input, the input function should specify what transformation needs to be done. In this case, the input function would be one that replaces all instances of a character by spaces. As it turns out the gsub() function does just that.
Here is the R code to build the content transformer, which we will call toSpace:
Now we can use this content transformer to eliminate colons and hypens like so:
docs <- tm_map(docs, toSpace, “:”)
Inspect random sections f corpus to check that the result is what you intend (use writeLines as shown earlier). To reiterate something I mentioned in the preamble, it is good practice to inspect the a subset of the corpus after each transformation.
If it all looks good, we can now apply the removePunctuation transformation. This is done as follows:
Inspecting the corpus reveals that several “non-standard” punctuation marks have not been removed. These include the single curly quote marks and a space-hyphen combination. These can be removed using our custom content transformer, toSpace. Note that you might want to copy-n-paste these symbols directly from the relevant text file to ensure that they are accurately represented in toSpace.
docs <- tm_map(docs, toSpace, “‘”)
docs <- tm_map(docs, toSpace, ” -“)
Inspect the corpus again to ensure that the offenders have been eliminated. This is also a good time to check for any other special symbols that may need to be removed manually.
If all is well, you can move to the next step which is to:
- Convert the corpus to lower case
- Remove all numbers.
Since R is case sensitive, “Text” is not equal to “text” – and hence the rationale for converting to a standard case. However, although there is a tolower transformation, it is not a part of the standard tm transformations (see the output of getTransformations() in the previous section). For this reason, we have to convert tolower into a transformation that can handle a corpus object properly. This is done with the help of our new friend, content_transformer.
Here’s the relevant code:
docs <- tm_map(docs,content_transformer(tolower))
Text analysts are typically not interested in numbers since these do not usually contribute to the meaning of the text. However, this may not always be so. For example, it is definitely not the case if one is interested in getting a count of the number of times a particular year appears in a corpus. This does not need to be wrapped in content_transformer as it is a standard transformation in tm.
docs <- tm_map(docs, removeNumbers)
Once again, be sure to inspect the corpus before proceeding.
The next step is to remove common words from the text. These include words such as articles (a, an, the), conjunctions (and, or but etc.), common verbs (is), qualifiers (yet, however etc) . The tm package includes a standard list of such stop words as they are referred to. We remove stop words using the standard removeWords transformation like so:
docs <- tm_map(docs, removeWords, stopwords(“english”))
Finally, we remove all extraneous whitespaces using the stripWhitespace transformation:
docs <- tm_map(docs, stripWhitespace)
Stemming
Typically a large corpus will contain many words that have a common root – for example: offer, offered and offering. Stemming is the process of reducing such related words to their common root, which in this case would be the word offer.
Simple stemming algorithms (such as the one in tm) are relatively crude: they work by chopping off the ends of words. This can cause problems: for example, the words mate and mating might be reduced to mat instead of mate. That said, the overall benefit gained from stemming more than makes up for the downside of such special cases.
To see what stemming does, let’s take a look at the last few lines of the corpus before and after stemming. Here’s what the last bit looks like prior to stemming (note that this may differ for you, depending on the ordering of the corpus source files in your directory):
flexibility eye beholder action increase organisational flexibility say redeploying employees likely seen affected move constrains individual flexibility dual meaning characteristic many organizational platitudes excellence synergy andgovernance interesting exercise analyse platitudes expose difference espoused actual meanings sign wishing many hours platitude deconstructing fun
Now let’s stem the corpus and reinspect it.
writeLines(as.character(docs[[30]]))
flexibl eye behold action increas organis flexibl say redeploy employe like seen affect move constrain individu flexibl dual mean characterist mani organiz platitud excel synergi andgovern interest exercis analys platitud expos differ espous actual mean sign wish mani hour platitud deconstruct fun
A careful comparison of the two paragraphs reveals the benefits and tradeoff of this relatively crude process.
There is a more sophisticated procedure called lemmatization that takes grammatical context into account. Among other things, determining the lemma of a word requires a knowledge of its part of speech (POS) – i.e. whether it is a noun, adjective etc. There are POS taggers that automate the process of tagging terms with their parts of speech. Although POS taggers are available for R (see this one, for example), I will not go into this topic here as it would make a long post even longer.
On another important note, the output of the corpus also shows up a problem or two. First, organiz and organis are actually variants of the same stem organ. Clearly, they should be merged. Second, the word andgovern should be separated out into and and govern (this is an error in the original text). These (and other errors of their ilk) can and should be fixed up before proceeding. This is easily done using gsub() wrapped in content_transformer. Here is the code to clean up these and a few other issues that I found:
Note that I have removed the stop words and and in in the 3rd and 4th transforms above.
There are definitely other errors that need to be cleaned up, but I’ll leave these for you to detect and remove.
The document term matrix
The next step in the process is the creation of the document term matrix (DTM)– a matrix that lists all occurrences of words in the corpus, by document. In the DTM, the documents are represented by rows and the terms (or words) by columns. If a word occurs in a particular document, then the matrix entry for corresponding to that row and column is 1, else it is 0 (multiple occurrences within a document are recorded – that is, if a word occurs twice in a document, it is recorded as “2” in the relevant matrix entry).
A simple example might serve to explain the structure of the TDM more clearly. Assume we have a simple corpus consisting of two documents, Doc1 and Doc2, with the following content:
Doc1: bananas are yellow
Doc2: bananas are good
The DTM for this corpus would look like:
| bananas | are | yellow | good | |
| Doc1 | 1 | 1 | 1 | 0 |
| Doc2 | 1 | 1 | 0 | 1 |
Clearly there is nothing special about rows and columns – we could just as easily transpose them. If we did so, we’d get a term document matrix (TDM) in which the terms are rows and documents columns. One can work with either a DTM or TDM. I’ll use the DTM in what follows.
There are a couple of general points worth making before we proceed. Firstly, DTMs (or TDMs) can be huge – the dimension of the matrix would be number of document x the number of words in the corpus. Secondly, it is clear that the large majority of words will appear only in a few documents. As a result a DTM is invariably sparse – that is, a large number of its entries are 0.
The business of creating a DTM (or TDM) in R is as simple as:
This creates a term document matrix from the corpus and stores the result in the variable dtm. One can get summary information on the matrix by typing the variable name in the console and hitting return:
<<DocumentTermMatrix (documents: 30, terms: 4209)>>
Non-/sparse entries: 14252/112018
Sparsity : 89%
Maximal term length: 48
Weighting : term frequency (tf)
This is a 30 x 4209 dimension matrix in which 89% of the rows are zero.
One can inspect the DTM, and you might want to do so for fun. However, it isn’t particularly illuminating because of the sheer volume of information that will flash up on the console. To limit the information displayed, one can inspect a small section of it like so:
<<DocumentTermMatrix (documents: 2, terms: 6)>>
Non-/sparse entries: 0/12
Sparsity : 100%
Maximal term length: 8
Weighting : term frequency (tf)
Docs creation creativ credibl credit crimin crinkl
BeyondEntitiesAndRelationships.txt 0 0 0 0 0 0
bigdata.txt 0 0 0 0 0 0
This command displays terms 1000 through 1005 in the first two rows of the DTM. Note that your results may differ.
Mining the corpus
Notice that in constructing the TDM, we have converted a corpus of text into a mathematical object that can be analysed using quantitative techniques of matrix algebra. It should be no surprise, therefore, that the TDM (or DTM) is the starting point for quantitative text analysis.
For example, to get the frequency of occurrence of each word in the corpus, we simply sum over all rows to give column sums:
Here we have first converted the TDM into a mathematical matrix using the as.matrix() function. We have then summed over all rows to give us the totals for each column (term). The result is stored in the (column matrix) variable freq.
Check that the dimension of freq equals the number of terms:
length(freq)
[1] 4209
Next, we sort freq in descending order of term count:
ord <- order(freq,decreasing=TRUE)
Then list the most and least frequently occurring terms:
freq[head(ord)]
one organ can manag work system
314 268 244 222 202 193
#inspect least frequently occurring terms
freq[tail(ord)]
yield yorkshir youtub zeno zero zulli
1 1 1 1 1 1
The least frequent terms can be more interesting than one might think. This is because terms that occur rarely are likely to be more descriptive of specific documents. Indeed, I can recall the posts in which I have referred to Yorkshire, Zeno’s Paradox and Mr. Lou Zulli without having to go back to the corpus, but I’d have a hard time enumerating the posts in which I’ve used the word system.
There are at least a couple of ways to simple ways to strike a balance between frequency and specificity. One way is to use so-called inverse document frequencies. A simpler approach is to eliminate words that occur in a large fraction of corpus documents. The latter addresses another issue that is evident in the above. We deal with this now.
Words like “can” and “one” give us no information about the subject matter of the documents in which they occur. They can therefore be eliminated without loss. Indeed, they ought to have been eliminated by the stopword removal we did earlier. However, since such words occur very frequently – virtually in all documents – we can remove them by enforcing bounds when creating the DTM, like so:
bounds = list(global = c(3,27))))
Here we have told R to include only those words that occur in 3 to 27 documents. We have also enforced lower and upper limit to length of the words included (between 4 and 20 characters).
Inspecting the new DTM:
<<DocumentTermMatrix (documents: 30, terms: 1290)>>
Non-/sparse entries: 10002/28698
Sparsity : 74%
Maximal term length: 15
Weighting : term frequency (tf)
The dimension is reduced to 30 x 1290.
Let’s calculate the cumulative frequencies of words across documents and sort as before:
#length should be total number of terms
length(freqr)
[1] 1290
#create sort order (asc)
ordr <- order(freqr,decreasing=TRUE)
#inspect most frequently occurring terms
freqr[head(ordr)]
organ manag work system project problem
268 222 202 193 184 171
#inspect least frequently occurring terms
freqr[tail(ordr)]
wait warehous welcom whiteboard wider widespread
3 3 3 3 3 3
The results make sense: the top 6 keywords are pretty good descriptors of what my blogs is about – projects, management and systems. However, not all high frequency words need be significant. What they do, is give you an idea of potential classification terms.
That done, let’s take get a list of terms that occur at least a 100 times in the entire corpus. This is easily done using the findFreqTerms() function as follows:
[1] “action” “approach” “base” “busi” “chang” “consult” “data” “decis” “design”
[10] “develop” “differ” “discuss” “enterpris” “exampl” “group” “howev” “import” “issu”
[19] “like” “make” “manag” “mani” “model” “often” “organ” “peopl” “point”
[28] “practic” “problem” “process” “project” “question” “said” “system” “thing” “think”
[37] “time” “understand” “view” “well” “will” “work”
Here I have asked findFreqTerms() to return all terms that occur more than 80 times in the entire corpus. Note, however, that the result is ordered alphabetically, not by frequency.
Now that we have the most frequently occurring terms in hand, we can check for correlations between some of these and other terms that occur in the corpus. In this context, correlation is a quantitative measure of the co-occurrence of words in multiple documents.
The tm package provides the findAssocs() function to do this. One needs to specify the DTM, the term of interest and the correlation limit. The latter is a number between 0 and 1 that serves as a lower bound for the strength of correlation between the search and result terms. For example, if the correlation limit is 1, findAssocs() will return only those words that always co-occur with the search term. A correlation limit of 0.5 will return terms that have a search term co-occurrence of at least 50% and so on.
Here are the results of running findAssocs() on some of the frequently occurring terms (system, project, organis) at a correlation of 60%.
project
inher 0.82
handl 0.68
manag 0.68
occurr 0.68
manager’ 0.66
enterpris
agil 0.80
realist 0.78
increment 0.77
upfront 0.77
technolog 0.70
neither 0.69
solv 0.69
adapt 0.67
architectur 0.67
happi 0.67
movement 0.67
architect 0.66
chanc 0.65
fine 0.64
featur 0.63
system
design 0.78
subset 0.78
adopt 0.77
user 0.77
involv 0.71
specifi 0.71
function 0.70
intend 0.67
softwar 0.67
step 0.67
compos 0.66
intent 0.66
specif 0.66
depart 0.65
phone 0.63
frequent 0.62
today 0.62
pattern 0.61
cognit 0.60
wherea 0.60
An important point to note that the presence of a term in these list is not indicative of its frequency. Rather it is a measure of the frequency with which the two (search and result term) co-occur (or show up together) in documents across . Note also, that it is not an indicator of nearness or contiguity. Indeed, it cannot be because the document term matrix does not store any information on proximity of terms, it is simply a “bag of words.”
That said, one can already see that the correlations throw up interesting combinations – for example, project and manag, or enterpris and agil or architect/architecture, or system and design or adopt. These give one further insights into potential classifications.
As it turned out, the very basic techniques listed above were enough for me to get a handle on the original problem that led me to text mining – the analysis of free text problem descriptions in my organisation’s service management tool. What I did was to work my way through the top 50 terms and find their associations. These revealed a number of sets of keywords that occurred in multiple problem descriptions, which was good enough for me to define some useful sub-categories. These are currently being reviewed by the service management team. While they’re busy with that that, I’m looking into refining these further using techniques such as cluster analysis and tokenization. A simple case of the latter would be to look at two-word combinations in the text (technically referred to as bigrams). As one might imagine, the dimensionality of the DTM will quickly get out of hand as one considers larger multi-word combinations.
Anyway, all that and more will topics have to wait for future articles as this piece is much too long already. That said, there is one thing I absolutely must touch upon before signing off. Do stay, I think you’ll find it interesting.
Basic graphics
One of the really cool things about R is its graphing capability. I’ll do just a couple of simple examples to give you a flavour of its power and cool factor. There are lots of nice examples on the Web that you can try out for yourself.
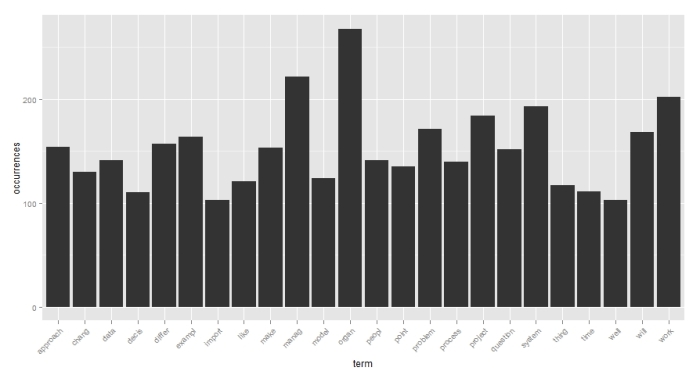
Let’s first do a simple frequency histogram. I’ll use the ggplot2 package, written by Hadley Wickham to do this. Here’s the code:
library(ggplot2)
p <- ggplot(subset(wf, freqr>100), aes(term, occurrences))
p <- p + geom_bar(stat=”identity”)
p <- p + theme(axis.text.x=element_text(angle=45, hjust=1))
p
Figure 1 shows the result.
The first line creates a data frame – a list of columns of equal length. A data frame also contains the name of the columns – in this case these are term and occurrence respectively. We then invoke ggplot(), telling it to consider plot only those terms that occur more than 100 times. The aes option in ggplot describes plot aesthetics – in this case, we use it to specify the x and y axis labels. The stat=”identity” option in geom_bar () ensures that the height of each bar is proportional to the data value that is mapped to the y-axis (i.e occurrences). The last line specifies that the x-axis labels should be at a 45 degree angle and should be horizontally justified (see what happens if you leave this out). Check out the voluminous ggplot documentation for more or better yet, this quick introduction to ggplot2 by Edwin Chen.
Finally, let’s create a wordcloud for no other reason than everyone who can seems to be doing it. The code for this is:
library(wordcloud)
#setting the same seed each time ensures consistent look across clouds
set.seed(42)
#limit words by specifying min frequency
wordcloud(names(freqr),freqr, min.freq=70)
The result is shown Figure 2.
Here we first load the wordcloud package which is not loaded by default. Setting a seed number ensures that you get the same look each time (try running it without setting a seed). The arguments of the wordcloud() function are straightforward enough. Note that one can specify the maximum number of words to be included instead of the minimum frequency (as I have done above). See the word cloud documentation for more.
This word cloud also makes it clear that stop word removal has not done its job well, there are a number of words it has missed (also and however, for example). These can be removed by augmenting the built-in stop word list with a custom one. This is left as an exercise for the reader :-).
Finally, one can make the wordcloud more visually appealing by adding colour as follows:
wordcloud(names(freqr),freqr,min.freq=70,colors=brewer.pal(6,”Dark2″))
The result is shown Figure 3.
You may need to load the RColorBrewer package to get this to work. Check out the brewer documentation to experiment with more colouring options.
Wrapping up
This brings me to the end of this rather long (but I hope, comprehensible) introduction to text mining R. It should be clear that despite the length of the article, I’ve covered only the most rudimentary basics. Nevertheless, I hope I’ve succeeded in conveying a sense of the possibilities in the vast and rapidly-expanding discipline of text analytics.
Note added on July 22nd, 2015:
If you liked this piece, you may want to check out the sequel – a gentle introduction to cluster analysis using R.
Note added on September 29th 2015:
…and my introductory piece on topic modeling.
Note added on December 3rd 2015:
…and my article on visualizing relationships between documents.




Reblogged this on Top GP and commented:
Excelente tutorial básico de como realizar text-mining (mineração de textos) usando a linguagem R.
LikeLiked by 1 person
Marco Alan Rotta
June 9, 2015 at 4:24 am
Nice tutorial. I recently posted a blog about how to use R to pull tweets.
https://arpitsolanki14.wordpress.com/2017/10/19/fetch-tweets-using-twitter-package-in-r/
LikeLiked by 1 person
arpitsolankiblog
October 20, 2017 at 2:32 am
[…] second part of my introductory series on text analysis using R (the first article can be accessed here). My aim in the present piece is to provide a practical introduction to cluster analysis. […]
LikeLike
A gentle introduction to cluster analysis using R | Eight to Late
July 22, 2015 at 8:53 pm
[…] that’s right, for free! All you need to is the open-source statistical package R (check out this section of my article on text mining for more on installing and using R) and the willingness to roll-up […]
LikeLike
Setting up an internal data analytics practice – some thoughts from a wayfarer | Eight to Late
September 3, 2015 at 8:28 pm
thanks! very useful and clear!
quick note:
perhaps
reqr <- colSums(as.matrix(dtmr))
#length should be total number of terms
length(freqr)
should start with
freqr <- colSums(as.matrix(dtmr))
Keep up the excellent work.
LikeLiked by 1 person
fernandoloizides
September 17, 2015 at 8:59 pm
Hi Fernando,
Many thanks for your feedback and for catching the error (fixed now).
Regards,
Kailash.
LikeLike
K
September 17, 2015 at 9:07 pm
[…] example which uses the topicmodels library in R. As in my previous articles in this series (see this post and this one), I will discuss the steps in detail along with explanations and provide […]
LikeLike
A gentle introduction to topic modeling using R | Eight to Late
September 29, 2015 at 7:18 pm
I would like to pass on a little tip regarding the tm package. Suppose you have a term-document matrix (terms=rows, documents=columns) named tdm. There are 3 vectors for your tdm object that are quite useful; tdm$i, tdm$j, tdm$v. tdm$i has, at each position, the term number in your term-document matrix. tdm$j contains the document number at that position. tdm$v contains the frequency of that term in that document. Suppose you want to know which documents term number 13 appears in, the following command will tell you:
>tdm$j[which(tdm$i==13)]
To find out the frequency of term 13 in each document, use the following command:
>tdm$v[tdm$j[which(tdm$i==13)]]
I found this quite useful for quick, ad hoc results on specific terms I was interested in. Hope you may find this useful
LikeLiked by 3 people
Charles Howard
October 8, 2015 at 11:29 pm
[…] I’m going to assume you have R and RStudio installed on your computer. If you need help with this, please follow the instructions here. […]
LikeLike
A gentle introduction to Naïve Bayes classification using R | Eight to Late
November 6, 2015 at 7:34 am
Muy buen tutorial…me encuentro haciendo un proyecto de text mining y tu información me ayudado mucho…cualquier inquietud te la haré llegar y espero me ayudes.
LikeLiked by 1 person
Robert
November 6, 2015 at 11:13 pm
Hi Robert,
I’m glad you found the article useful. Thanks for reading and commenting!
Regards,
Kailash.
LikeLike
K
November 7, 2015 at 8:39 am
When I downloaded your text files your single curly came in to my .txt as í. So I’ll is Iíll and don’t is donít in TheDilemmasOfEnterprise.txt. (26) In R Studio they come up as 92.
What symbol do I enter into this line of code: docs <- tm_map(docs, toSpace, “’”)
Are there any other symbol modifications I should check for in this step?
thanks
LikeLiked by 1 person
Stav
November 18, 2015 at 12:39 am
Hi Stav,
Thanks for reading and trying out the code. Yes, this is a problem from time to time. The quick and dirty way that usually works is to cut and paste the exact symbol from the textf ile into the relevant tm_map argument. As for other symbols, I removed them as I found them, but I haven’t maintained a list.
Also, note that the 1) tm functions do not always work as advertised and 2) the order of preprocessing steps can make a difference (this point has been noted by others as well).
Hope this helps.
Regards,
Kailash.
LikeLiked by 1 person
K
November 18, 2015 at 7:34 am
[…] of documents, a process which I have dealt with at length in my introductory pieces on text mining and topic modeling. In fact, the steps are actually identical to those detailed in the second […]
LikeLike
A gentle introduction to network graphs using R and Gephi | Eight to Late
December 2, 2015 at 7:20 am
Thank you so much! I have been working on my script for two days now and this was such a wonderful help! I would like to note that my R did not like your ” “, I have to change the ” ” when I wrote my script. It might because of the font changing over or something? The point being is that I had to change the ” ” manually in order for R to process properly otherwise R just gave me error messages. Hopefully no one else ran into that problem! Thank you so much!!! 🙂
LikeLiked by 1 person
Kurt
December 12, 2015 at 5:14 am
This is an awesome tutorial! Thanks. Keep up the good work.
LikeLiked by 1 person
Fidelis
December 23, 2015 at 11:05 am
[…] Preamble This article is based on my exploration of the basic text mining capabilities of R, the open source statistical software. It is intended primarily as a tutorial for novices in text mining as well as R. […]
LikeLike
A gentle introduction to text mining using R | ...
December 23, 2015 at 4:51 pm
[…] follows, I will use the open source software, R. If you are new to R, you may want to follow this link for more on the basics of setting up and installing it. Note that the R implementation of the […]
LikeLike
A gentle introduction to decision trees using R | Eight to Late
February 16, 2016 at 6:33 pm
Hi,
Really nice tutorial. In the preprocessing step, i am not able to remove bullet point. Please suggest what can be done.
Thank you
LikeLike
Appurv
February 19, 2016 at 8:38 pm
Hi,
Do you know how much your text is reduced via stemming? (I found a length reduction of about 14% in one of my text mining process)
As I write here (in french) : http://data-bzh.fr/text-mining-r-part-3/, I tend to think that stemming is not the best solution for visualisation purpose, as the words become less readable.
Anyway, great article 🙂
Best,
Colin
LikeLiked by 1 person
Colin Data Bzh
May 3, 2016 at 3:21 am
Hi, there! Just a tip for removing some special characters like “$” or “(” using the ‘toSpace’ content transformer: Change the function to:
wedopages
June 3, 2016 at 11:22 am
Great! Thanks for the tip.
Regards,
K.
LikeLike
K
June 22, 2016 at 10:53 am
[…] A gentle introduction to text mining using R […]
LikeLike
R프로그래밍 참고할 만한 사이트 | This Is YNWA
June 19, 2016 at 4:05 pm
Reblogged this on evolvingprogrammer and commented:
An excellent introduction to text mining in R. Very well explained.
LikeLike
evolvingprogrammer
June 30, 2016 at 7:26 am
Great tutorial!! I don’t comment much on tutorials I have used but this one really sorted me out…quite gentle for beginners…keep sharing your knowledge..
LikeLike
Peninah Njoka
July 8, 2016 at 9:41 pm
Thank you!
LikeLike
K
July 12, 2016 at 6:21 am
Awesome explanation. Great article sir!!
LikeLike
Subha
August 31, 2016 at 4:42 pm
This is excellent tutorial.. explained in detail!! If you could also include how to combine two or three words and find their frequency would be great.
LikeLike
SAF
September 9, 2016 at 12:27 am
[…] A gentle introduction to text mining using R | Eight to Late […]
LikeLike
daily 09/17/2016 | Cshonea's Blog
September 18, 2016 at 6:31 am
Good explanation…I would like use the term document matrix for regression analysis…for instance a logistic model…how could I do?
Thanks…
LikeLike
miguel
October 14, 2016 at 4:31 am
[…] further detail, check out Kailash Awati’s Gentle Introduction to Text Mining with R here and an RStudio resource here which describe how to text mine with R’s tm package. The […]
LikeLike
Text Mining with ‘tm’ | pjvwebb
October 27, 2016 at 5:49 pm
[…] When you want to learn textmining from someone who is (much) more knowledgeable than me: https://eight2late.wordpress.com/2015/05/27/a-gentle-introduction-to-text-mining-using-r […]
LikeLike
Getting started with (text-mining in) R – incurableRchiver
December 31, 2016 at 2:15 am
[…] I assume you have R and RStudio installed. For instructions on how to do this, have a look at the first article in this series. The processing preliminaries – loading libraries, data and creating training and test datasets […]
LikeLike
A gentle introduction to support vector machines using R | Eight to Late
February 7, 2017 at 8:27 pm
[…] one’s head around, the basics are simple enough. Yea, I really mean that – for proof, check out my tutorial on the […]
LikeLike
A prelude to machine learning | Eight to Late
February 23, 2017 at 3:13 pm
Hi I got an error message:
> docs <- Corpus(DirSource(“C:/Users/oliver/Documents/TextMining”))
Error: unexpected input in "docs <- Corpus(DirSource(“"
Do you mind to tell me what I did wrong. Thank you very much.
LikeLiked by 1 person
Oliver Chen
March 3, 2017 at 2:06 am
I got this too! Did you get any reply yet?
LikeLike
Amy P Case
November 14, 2017 at 11:45 am
Try retyping code directly into RStudio instead of a copy / paste. I suspect this happens because the quotes enclosing the path are rendered incorrectly by html.
Hope this helps.
Regards,
Kailash.
LikeLike
K
November 14, 2017 at 11:54 am
[…] Awati, Kailash. “A gentle introduction to text mining using R.” Eight to Late. Accessed March 16, 2017. https://eight2late.wordpress.com/2015/05/27/a-gentle-introduction-to-text-mining-using-r/. […]
LikeLike
Holy Indexes – Small Words Big Numbers
March 17, 2017 at 1:10 pm
Hands down on of the best tutorials out there. Thanks a ton
LikeLike
Sowhardh
April 7, 2017 at 6:04 am
This was a very helpful walkthrough. Bravo!
LikeLike
rk
April 11, 2017 at 2:31 am
Great tutorial. Thank you so much! 🙂
LikeLike
Georgi
May 3, 2017 at 11:01 pm
Dear K, This article is phenomenal. Thank you very much for posting this. This will help me to analyze the problem on my hand much better.
LikeLike
Venkataramanan T S
May 30, 2017 at 8:30 pm
Nice article K. Thanks
LikeLike
Vadivelan
June 2, 2017 at 9:03 pm
[…] range of enterprise and other software tools, including the popular (and free) R programming language, are available to implement text analytics. You can also visualize the results of text analytics […]
LikeLike
Words with (Association) Friends - Association Analytics
June 9, 2017 at 2:40 am
[…] of all, you would need a term document matrix (e.g. the one created by the tm package). See this piece on how to do that. With that, training an LDA model is as simple […]
LikeLike
Classifying documents into topics using LDA – Holla
June 12, 2017 at 6:55 pm
Thanks for the great tutorial!
LikeLike
Georgi
June 16, 2017 at 5:13 am
thank you its really useful
LikeLike
Deepa
October 2, 2017 at 4:42 pm
GRACIAS!!!! Thanks a lot! God bless you!
LikeLike
Maria Vasquez
October 4, 2017 at 3:48 am
Great Article. Could you please share if you any other similar article that you have written on Text Mining.
LikeLike
Hitesh
October 17, 2017 at 8:26 pm
Thanks Hitesh. Here’s one I wrote on visualising document similarity using R and Gephi: https://eight2late.wordpress.com/2015/12/02/a-gentle-introduction-to-network-graphs-using-r-and-gephi/
You can do the network graph easily enough using the igraph library instead of Gephi. I’ve done this for one of my classes but never got around to publishing it, but here’s an article to help you get started: https://www.r-bloggers.com/an-example-of-social-network-analysis-with-r-using-package-igraph/
Hope this helps.
Regards,
Kailash.
LikeLike
K
October 17, 2017 at 8:44 pm
Excellent tutorial on text mining. Perhaps you can point me in the right direction? My goal is to write a R script that takes a paragraph of text and identifies Twitter keywords in the text. In addition I would like to rank the keywords in the text by popularity on Twitter. Thanks, in advance.
LikeLike
Frank Cohen
October 25, 2017 at 12:09 am
This is really easy to follow, but I got stumped at this step:
> getwd()
[1] “C:/Users/caseap/Documents/TextMining”
> docs <- Corpus(DirSource(“C:/Users/caseap/Documents/TextMining”))
Error: unexpected input in "docs <- Corpus(DirSource(“"
Help!!
LikeLike
Amy P Case
November 14, 2017 at 11:47 am
See response above.
Regards,
Kailash.
LikeLiked by 1 person
K
November 14, 2017 at 11:54 am
Well that worked! But sadly, now I get this:
> docs <-Corpus(DirSource("C:/Users/caseap/Documents/TextMining"))
Error in Corpus(DirSource("C:/Users/caseap/Documents/TextMining")) :
could not find function "Corpus"
I installed Corpus (I think it was already installed as a dependency) again after getting this once, but same result.
Thank you for your easy-to-follow instructions! I really do need this level of explanation (even the "stating the obvious).
LikeLike
Amy P Case
November 15, 2017 at 3:18 am
You’ll need to load the tm package before calling any of its functions. The following line will do this:
library(tm)
LikeLike
K
November 15, 2017 at 11:13 am
I did that, too.
LikeLike
Amy P Case
November 15, 2017 at 11:32 am
Hi, I’m trying to follow your tutorial but I can’t remove the space-hyphen combinations even when I paste the correct hyphen directly from the actual text. Does anyone have any suggestions?
LikeLike
Nathan
November 15, 2017 at 2:39 am
Sorted it, just had to use ” \u2013″ and not ” –”
LikeLike
Nathan
November 15, 2017 at 3:20 am
[…] removed everything except my answers and organized the answers by day. I then found some guides for Text Mining in R and went to town. Each day’s answers were contained in separate text files, and each file […]
LikeLike
Heaps of Sheeps: stress, sleeplessness, and readability scores – Joshua D. Sites
November 18, 2017 at 10:20 am
Hey. Thanks for this extremely useful and comprehensive tutorial 🙂
I’m struggling with some strange output words in my topics/ggplot/wordcloud.
There sometimes is an “” at the end of some words. It seems as if it has problems with “ö,ä,ü…” or with “ß” which stands for double “s” in german.
Do you have any idea how I could get rid of this problem (of having  in the output)?
LikeLike
Alessio
December 14, 2017 at 7:23 pm
Hi Alessio,
This is likely an encoding issue. You could try using iconv as suggested in this article: https://stackoverflow.com/questions/18153504/removing-non-english-text-from-corpus-in-r-using-tm
Regards,
Kailash.
LikeLike
K
December 15, 2017 at 8:50 am
Thanks heaps!! Great article for beginners. In the DTM step though, I’m getting below error
Error in simple_triplet_matrix(i, j, v, nrow = length(terms), ncol = length(corpus), :
‘i, j’ invalid
Any idea what’s wrong?
LikeLike
rahulsinha7
October 8, 2018 at 4:08 pm
Found out, it was this command that was causing error
docs <- tm_map(docs, PlainTextDocument)
LikeLiked by 1 person
rahulsinha7
October 8, 2018 at 4:20 pm
Wow! This is excellent. Thanks for putting all of this together!
LikeLike
Niels Smith
November 22, 2018 at 2:49 am
Thanks for the feedback, glad you found it useful!
LikeLike
K
November 22, 2018 at 5:47 am
Hi..I’m working on a similar project that you initially mentioned in your intro..trying to analyse the ticket content for a technical support team. I’m new to R and i would like to know if this would work for an excel spreadsheet rather than a .text file? The data i have is very primitive and I would like to provide insights into the different types of tickets ,key words used in the subject and description etc
LikeLike
Meg
November 27, 2018 at 11:20 am
Hi Meg,
Yes, you can load up data from an Excel spreadsheet or csv into a dataframe and then create a corpus from the dataframe. Here’s the code to do that:
library(tm)
#read dataset stored in text.csv
#assuming csv file has a header and possibly contains multiple columns
text_dataframe <- read.csv(file="text_data.csv",header = T)
#Create corpus – assumes text data is in a column named "text_col"
text_corpus <- VCorpus(VectorSource(text_dataframe$text_col))
You would then proceed as discussed in the blog,
Hope this helps.
Regards,
Kailash.
LikeLike
K
November 29, 2018 at 10:04 am
Thank you
LikeLike
Meg
November 29, 2018 at 10:06 am
No worries. On a related note, if you’re interested in a practical introductory course on R / data science you may want to consider this:
http://datanitio.com
LikeLike
K
November 29, 2018 at 10:59 am
[…] book-reading habits / following the instructions / guide found here). A brief search revealed this ‘gentle introduction’, and this is what I’ll be more or less following in this series of […]
LikeLike
Text mining the Burney Corpus in R Part I. | Dan Waterfield
March 14, 2019 at 11:45 pm
Thank you for such a useful introduction! It’s first time trying out Text mining and you helped so much!
I have a small question for this code right here:
dtmr <-DocumentTermMatrix(docs, control=list(wordLengths=c(4, 20),
bounds = list(global = c(3,27))))
May I ask why you chose your global range to be 3 to 27. Is there a reason for it? And is there a recommended range for this when text mining? Or is it to my own discrete? Thank you
LikeLike
thelittlelatte
May 29, 2019 at 7:10 pm
These numbers are the lower and upper bounds for the number of documents in which a word appears. In effect, this tells tm to disregard words that occur in less than 3 or more than 27 documents. I’ve chosen these bounds because they seemed reasonable for a corpus with 30 docs. It’s up to you to judge what’s appropriate for your corpus,
Thanks for reading the article, I’m glad you found it useful.
Regards,
Kailash.
LikeLike
K
May 29, 2019 at 9:13 pm
[…] 对于英文菜名,我们使用tm包进行分析。这一部分的主要参考资料在这里。首先,我们将原数据中的英文菜名(NAME.ENG)单独提取出来,并将字符串拆成单词便于分析。由于我输入数据源时有提前考虑这个问题,所以英文菜名一律用小写字母,也没有缩写,所以数据算是比较干净的。 […]
LikeLike
Analysing FoodTruck Menu in R – II 用R分析餐车菜单(二) – Spark Tseung
June 7, 2019 at 6:39 am
Many thanks for the wonderful tutorial.
LikeLike
gaurav
February 1, 2020 at 4:23 pm
Hi, thanks for sharing your knowledge. I really enjoyed reading this. You have tried to explain everything so well. Keep up the good work.
LikeLike
Ranju
May 9, 2021 at 2:07 am
Thank you so much for this post. I have been doing statistical analyses using R and Stata, and have been trying to get into text analysis for my research. This was the best-explained intro tutorial into the subject matter that I could find. Thanks again-
LikeLike
Zeewan
May 19, 2021 at 5:07 pm